Collapse by node id
Introduction
COLLAPSE template are used to collapse branches.
Typically, The user merges branches A and B by folding them at their most recent common ancestor (MRCA). However, it is normally hard for users to declare the MRCA node directly. The node name is usually generated automatically by software rather than specified by the user when building the phylogenetic tree. Some programs do not even generate the node name.
This section has taken these complicated cases into account, and provide the corresponding solution, so that users can easily prepare the input data.
Node ID
Although we used a tip label in the above example, using node names directly is more convenient if it is available. Thus, the itol.toolkit also enable you to define the collapsed branch using the node name directly .
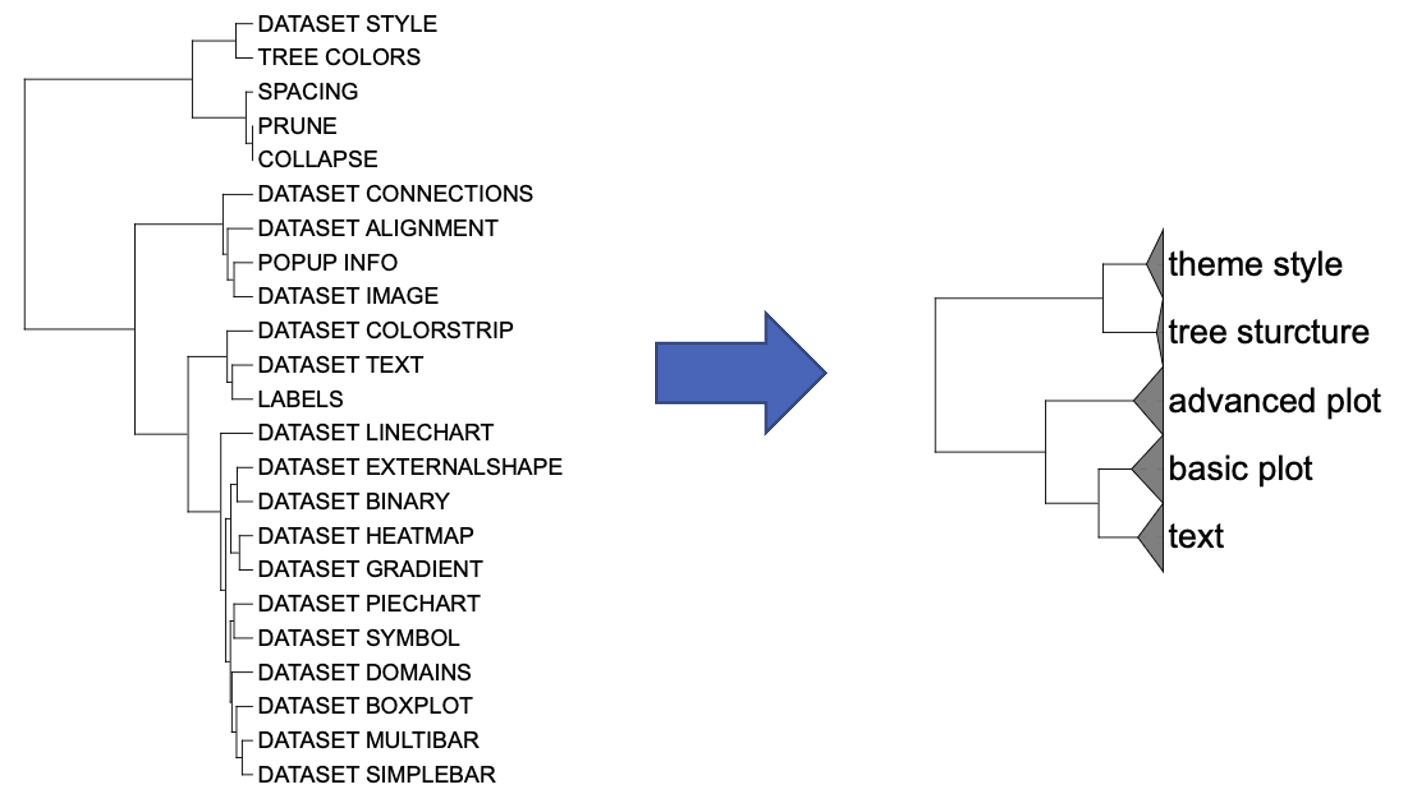
The tree provided by itol.toolkit dataset 1 including the node labels:
ape::read.tree(tree)
# Phylogenetic tree with 23 tips and 22 internal nodes.
#
# Tip labels:
# SPACING, COLLAPSE, PRUNE, TREE_COLORS, DATASET_STYLE, DATASET_CONNECTIONS, ...
# Node labels:
# I1, I2, tree_sturcture, I4, theme_style, I6, ...
#
# Rooted; includes branch lengths.
Therefore, you could specify the group you want to merge directly.
group_names <- unique(c("tree_sturcture", "theme_style", "text", "basic_plot"))
unit_2 <- create_unit(data = group_names,
key = "E002_collapse_2",
type = "COLLAPSE",
tree = tree)
write_unit(unit_2,
paste0(getwd(),"/E002_collapse_2.txt"))
If you have questions about how to upload results files from iTOL toolkit for interactive visualization on iTOL, please refer to the Operation document for step-by-step guidance.
# collapse