Text alignment & 20+ colors
DOI: 10.1016/j.jhazmat.2022.129230
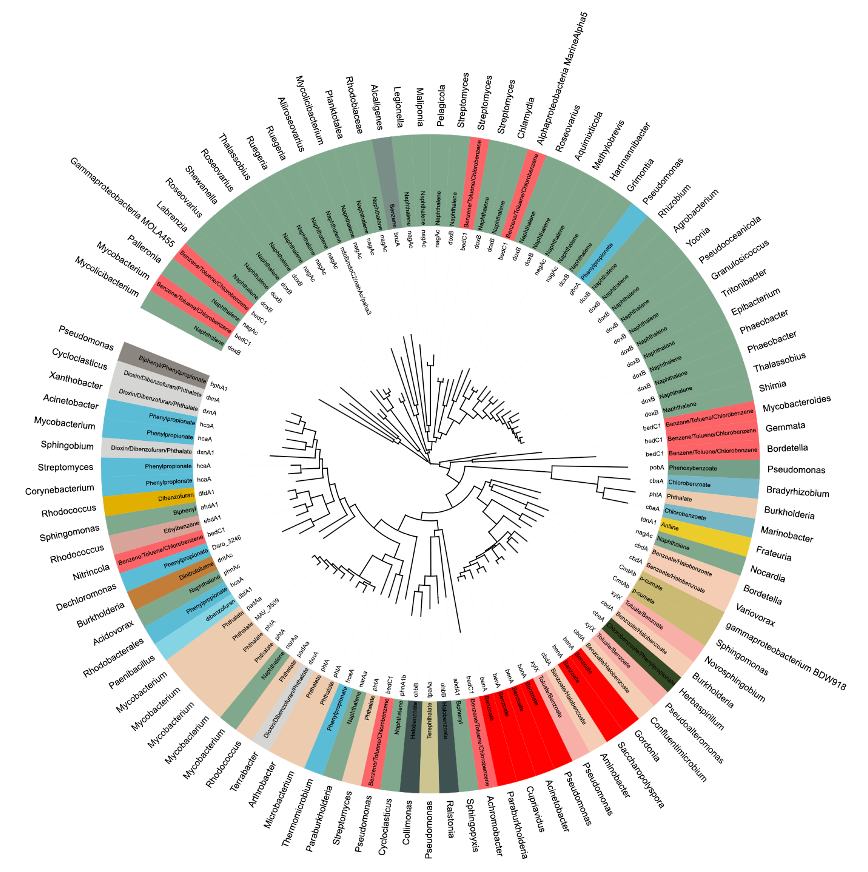
Figure 2
library(itol.toolkit)
library(data.table)
library(ape)
library(stringr)
tree <- "https://raw.githubusercontent.com/TongZhou2017/itol.toolkit/master/inst/extdata/dataset2/tree.nwk"
hub <- create_hub(tree)
data_file <- "https://raw.githubusercontent.com/TongZhou2017/itol.toolkit/master/inst/extdata/dataset2/metadata.txt"
data <- data.table::fread(data_file)
data <- data %>% filter(ID != "Seq30_")
select_tip = phylo$tip.label[phylo$tip.label != "Seq372"]
unit_1 <- create_unit(data = select_tip,
key = "rep_Li2022jhm_1_prune",
type = "PRUNE",
tree = tree)
df_gene <- data %>% select(ID, Gene_name)
unit_2 <- create_unit(data = df_gene,
key = "rep_Li2022jhm_4_text_1",
type = "DATASET_TEXT",
size_factor = 1,
rotation= 180,
position = -1,
color = "#000000",
tree = tree)
unit_2@common_themes$basic_theme$margin <- -500
df_relabel <- data %>% select(ID, Substrate)
unit_3 <- create_unit(data = df_relabel,
key = "rep_Li2022jhm_2_labels",
type = "LABELS",
tree = tree)
df_substrate <- data %>% select(ID, Substrate)
set.seed(666)
unit_4 <- create_unit(data = df_substrate,
key = "rep_Li2022jhm_3_range",
type = "TREE_COLORS",
subtype = "range",
color = "wesanderson",
tree = tree)
df_species <- data %>% select(ID, Species)
unit_5 <- create_unit(data = df_species,
key = "rep_Li2022jhm_5_text_2",
type = "DATASET_TEXT",
size_factor = 1,
rotation= 0,
position = -1,
color ="#000000",
tree = tree)
unit_5@common_themes$basic_theme$margin <- 50
hub <- hub + unit_1 + unit_2 + unit_3 + unit_4 + unit_5
write_hub(hub, getwd())