Load data
tab <- read.table("~/Downloads/ziyuzhang.txt",sep="\t",header=T,stringsAsFactors = FALSE, row.names = 1)
group <- as.data.frame(t(tab[1:2,]),stringsAsFactors =FALSE)
names(group) <- c("group","batch")
tab <- tab[-c(1,2),]
tab <- sapply(tab, as.numeric)
rownames(tab) <- 1:nrow(tab)
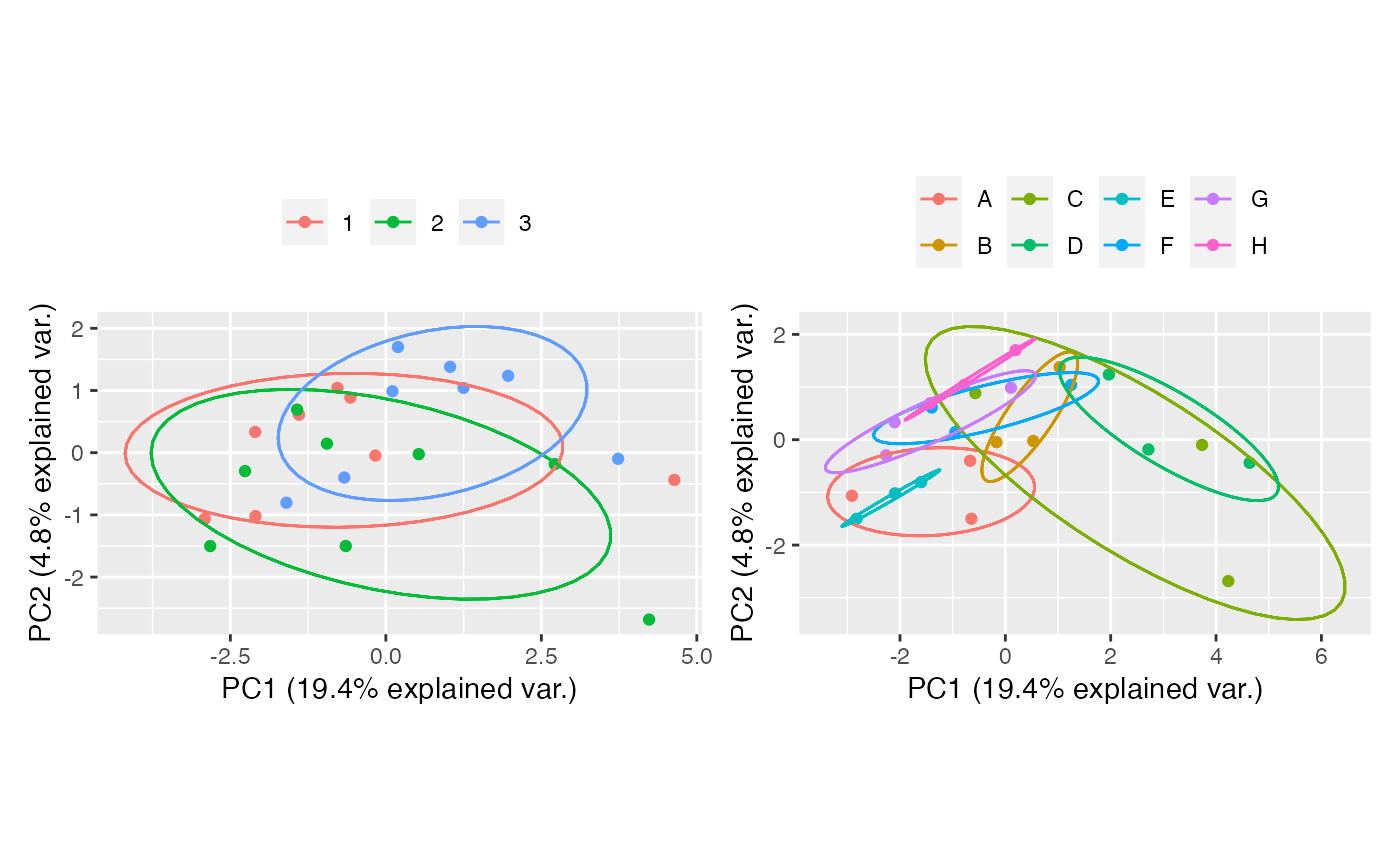
Raw
raw <- table_correct_batch(tab,"raw",plot = T,meta_data = group,batch = "batch",group = "group")
raw[[2]] + raw[[3]]
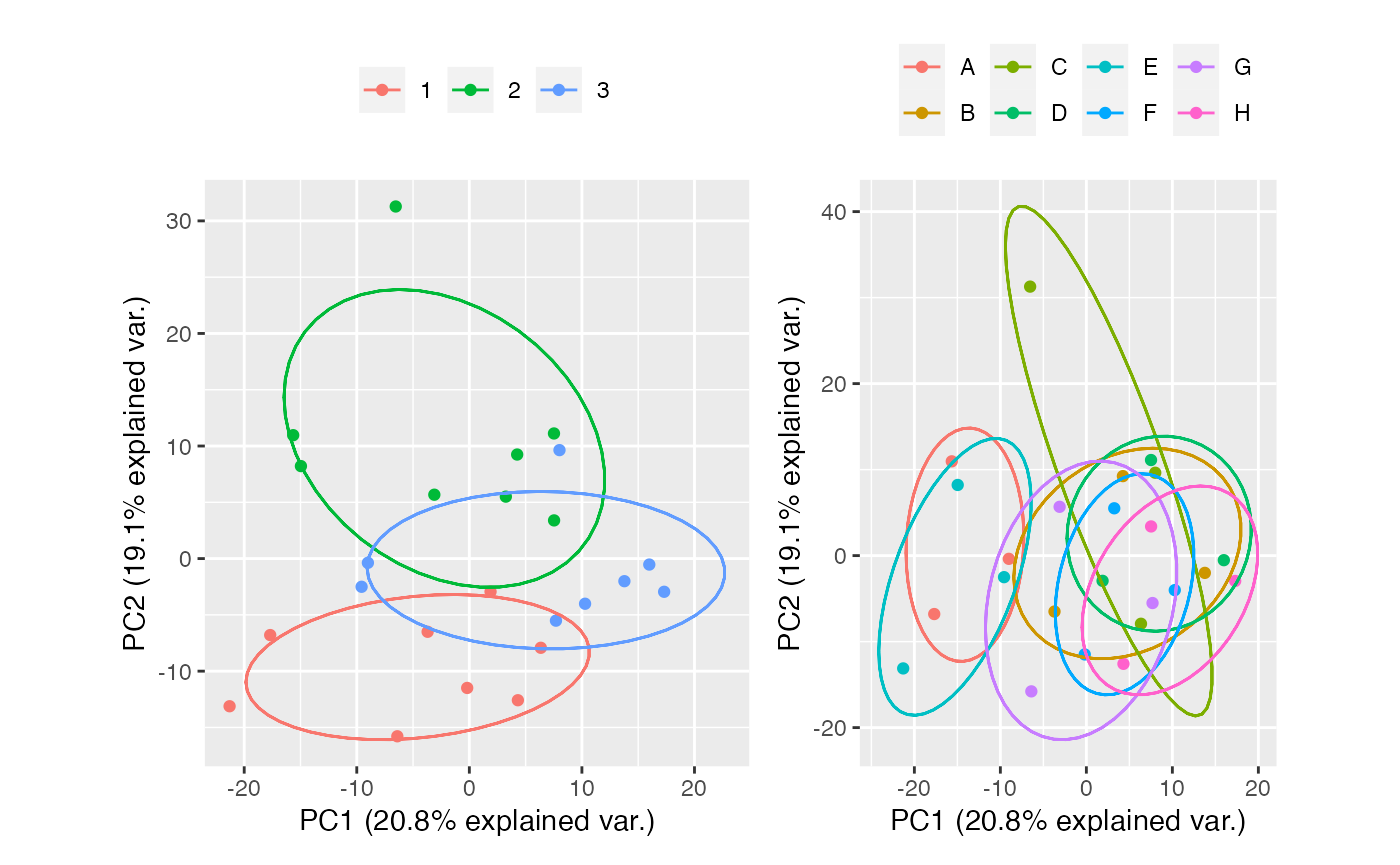
Combat
combat <- table_correct_batch(tab,"combat",plot = T,meta_data = group,batch = "batch",group = "group")
#> Standardizing Data across genes
combat[[2]] + combat[[3]]
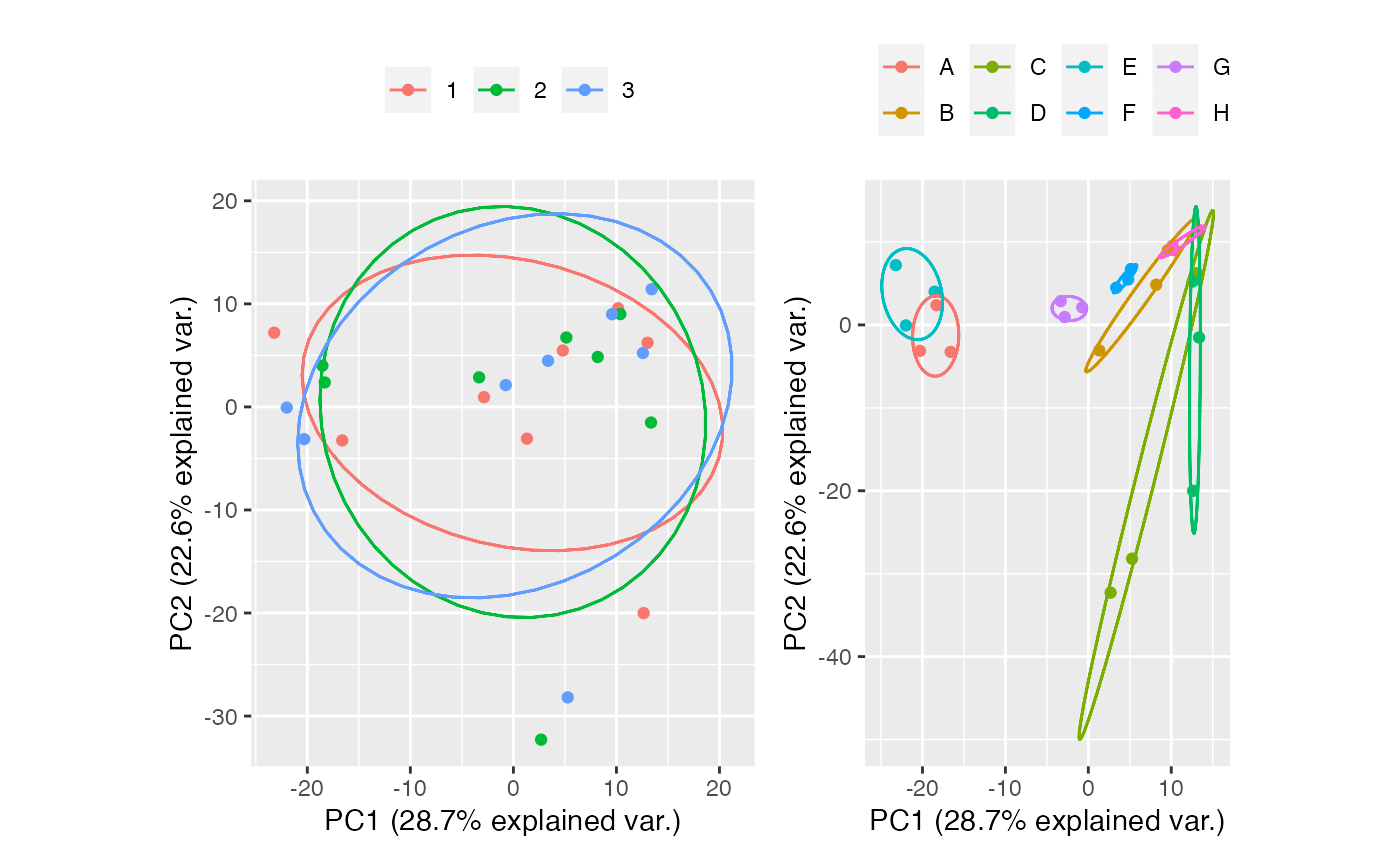
Limma
limma <- table_correct_batch(tab,"limma",plot = T,meta_data = group,batch = "batch",group = "group")
limma[[2]] + limma[[3]]
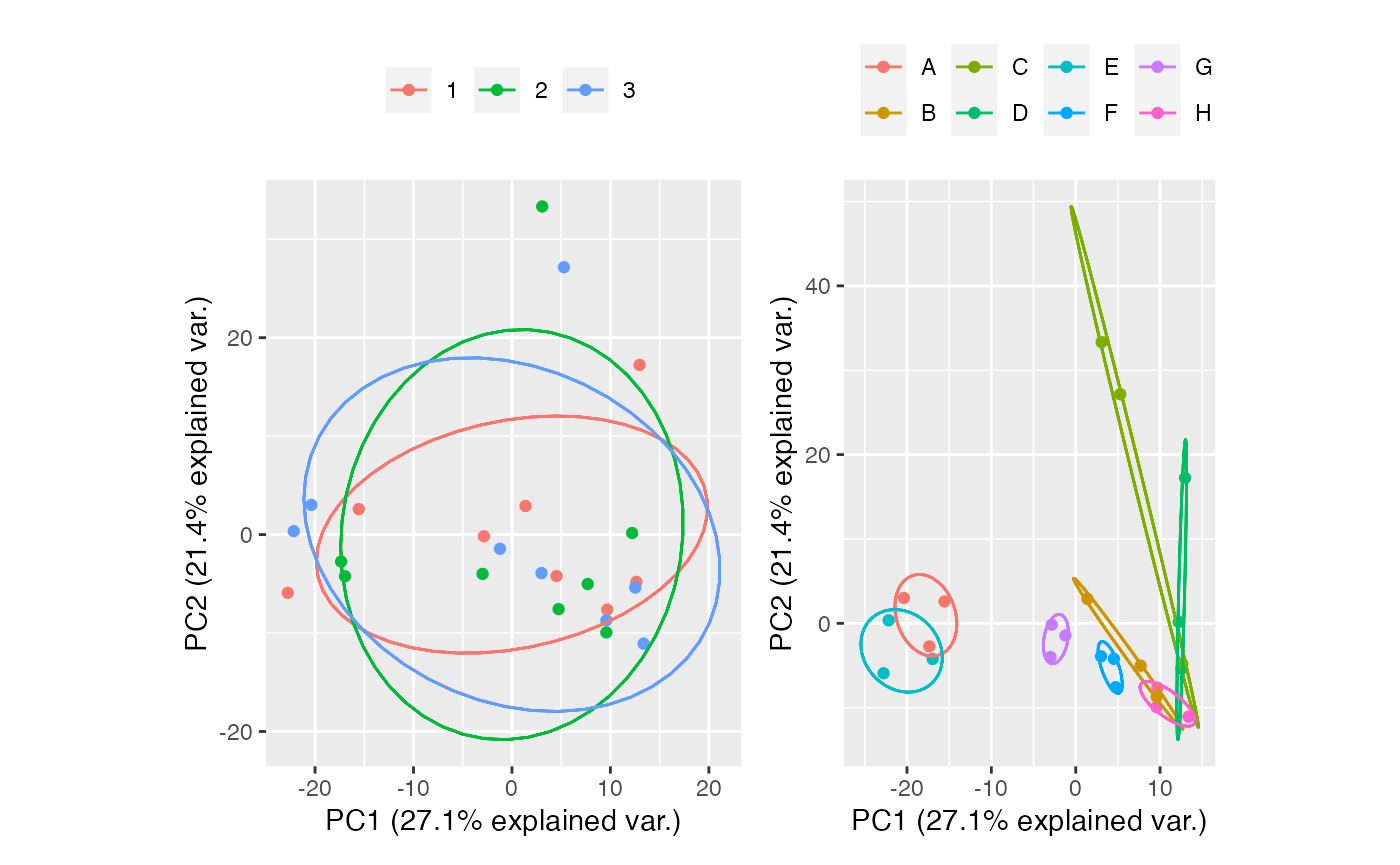
MNN
mnn <- table_correct_batch(tab,"mnn",plot = T,meta_data = group,batch = "batch",group = "group")
mnn[[2]] + mnn[[3]]
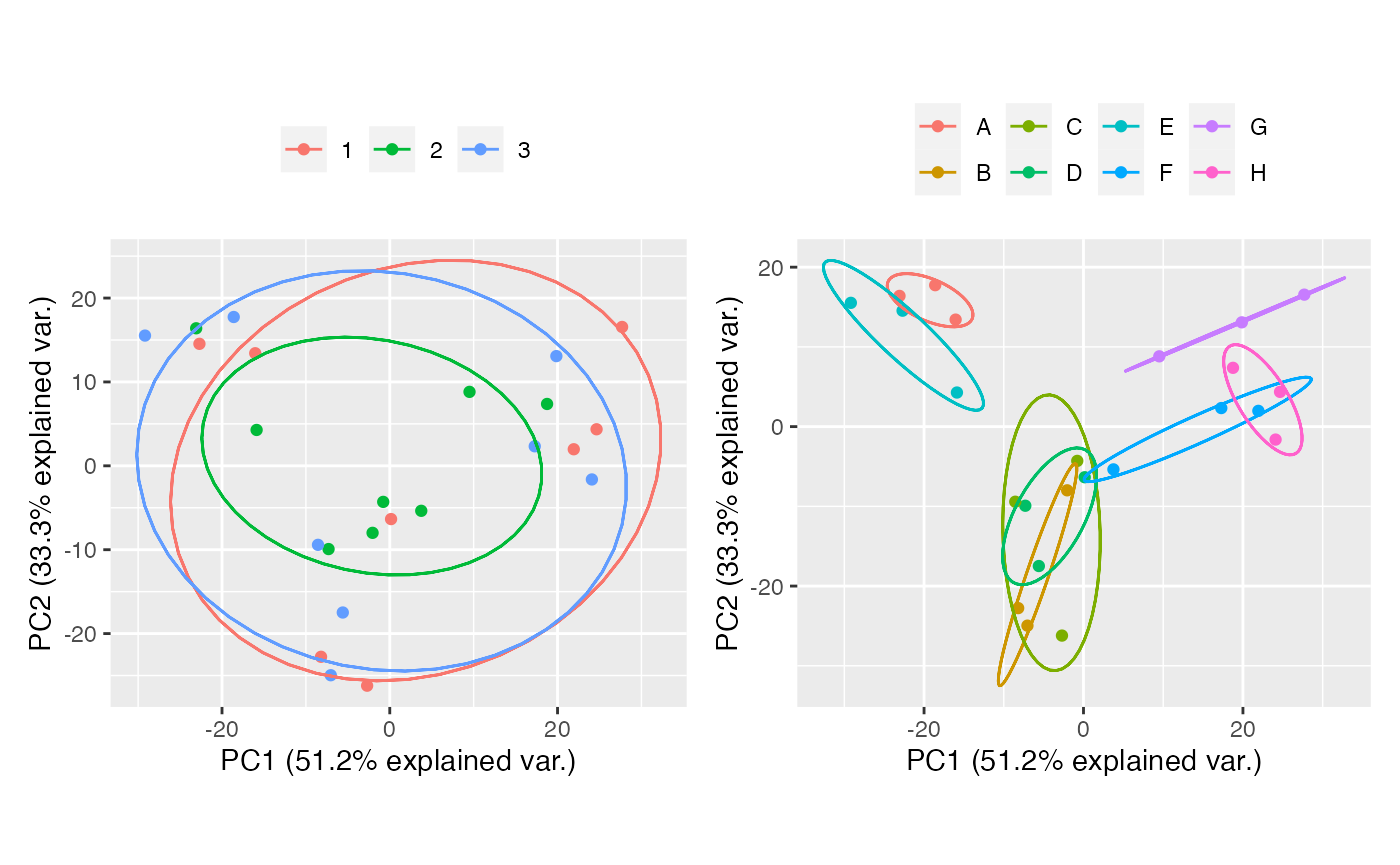
Harmony
harmony <- table_correct_batch(tab,"harmony",plot = T,meta_data = group,batch = "batch",group = "group")
harmony[[2]] + harmony[[3]]